{EcotaxaTools}
An R Package for Ecotaxa & Ecopart Processing. UVP User Workshop 2022
A Common Language

- Collaborative projects
- Common code for open science
- Ease-of-use
Current Configuration:
ver 1.2.0.9000
- Available on github
- Primarily focused on the UVP
- particularly built out for zooplankton processing
Package Basics
Package Basics with UVP
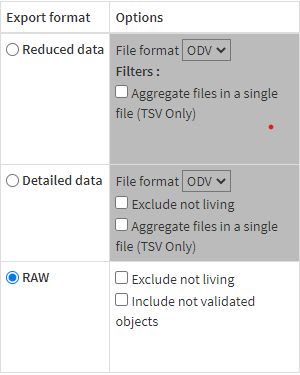
- Focus on raw-data export
- Files downloaded go in a directory
- A class/object structure to navigate R
Package Basics with UVP
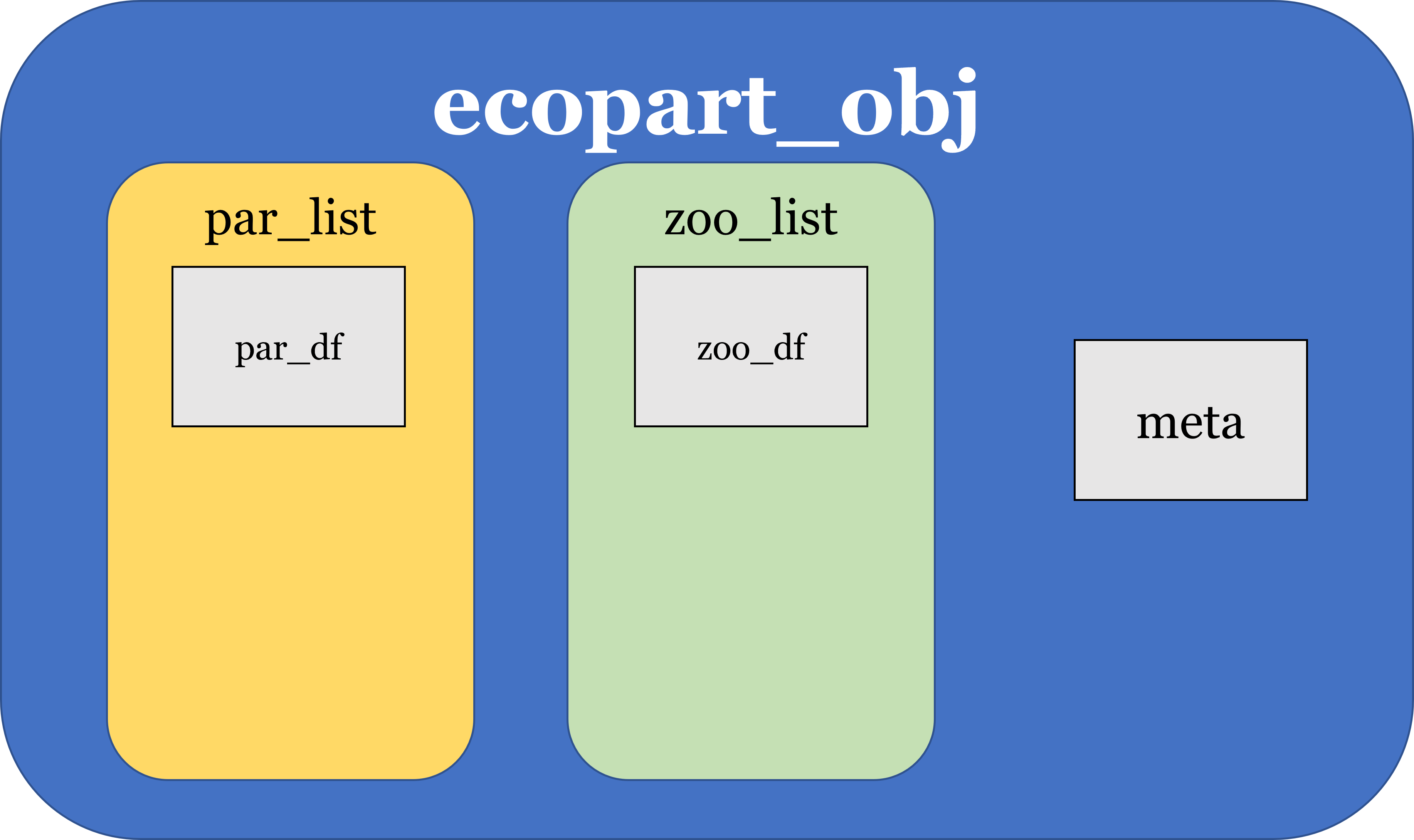
ecopart_obj class structure:
Package Basics with UVP
ecopart_obj class structure:
- Applying functions across all profiles
- Quickly summarize data
- Requires looping
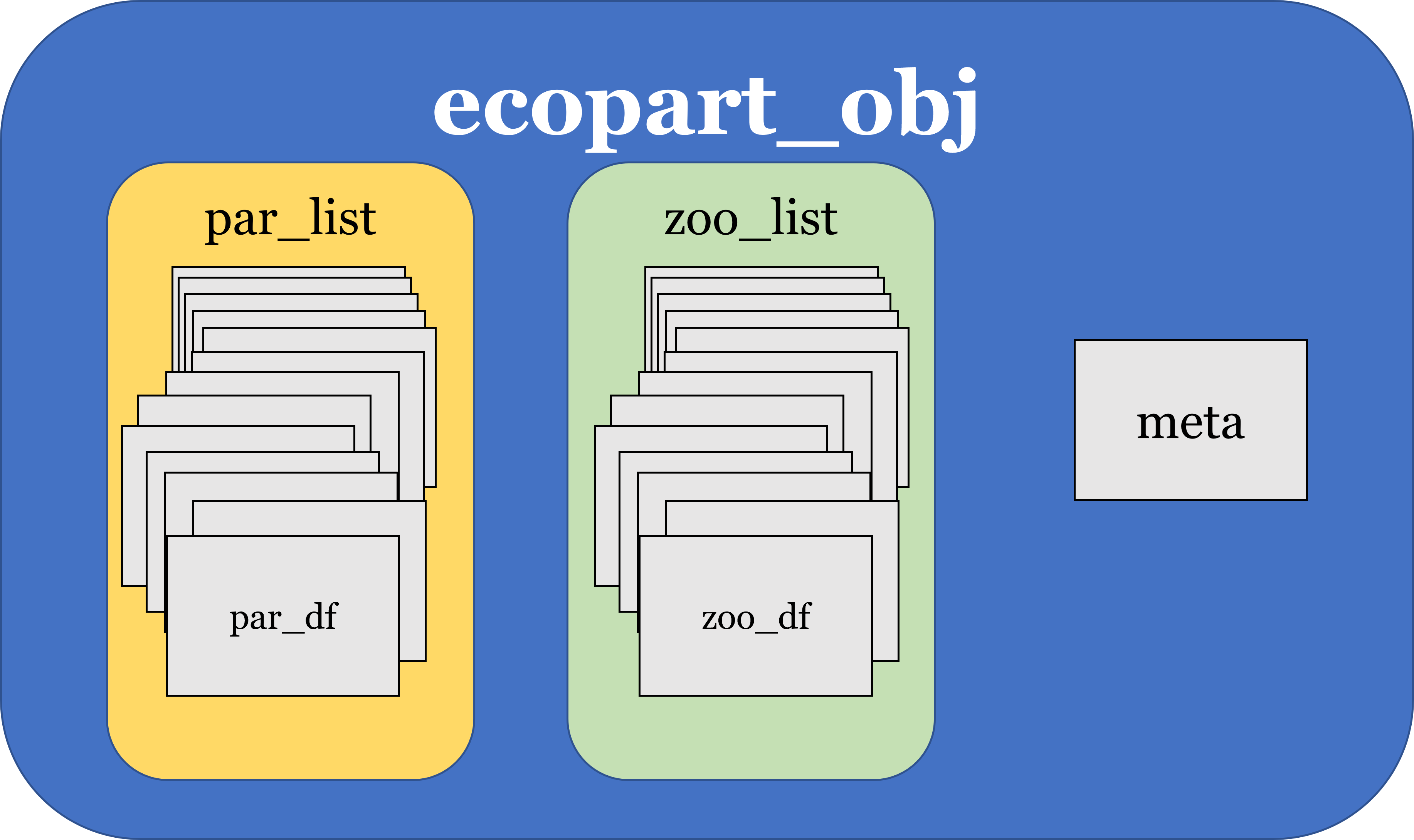
Package Basics with UVP
Code Considerations:
- Readability
- Minimalize dependancies
- Pipe friendly
|> - Rely on
lapply()

Package tools
Package tools
Project management:
Package tools
Project management:
- Quickly pull information
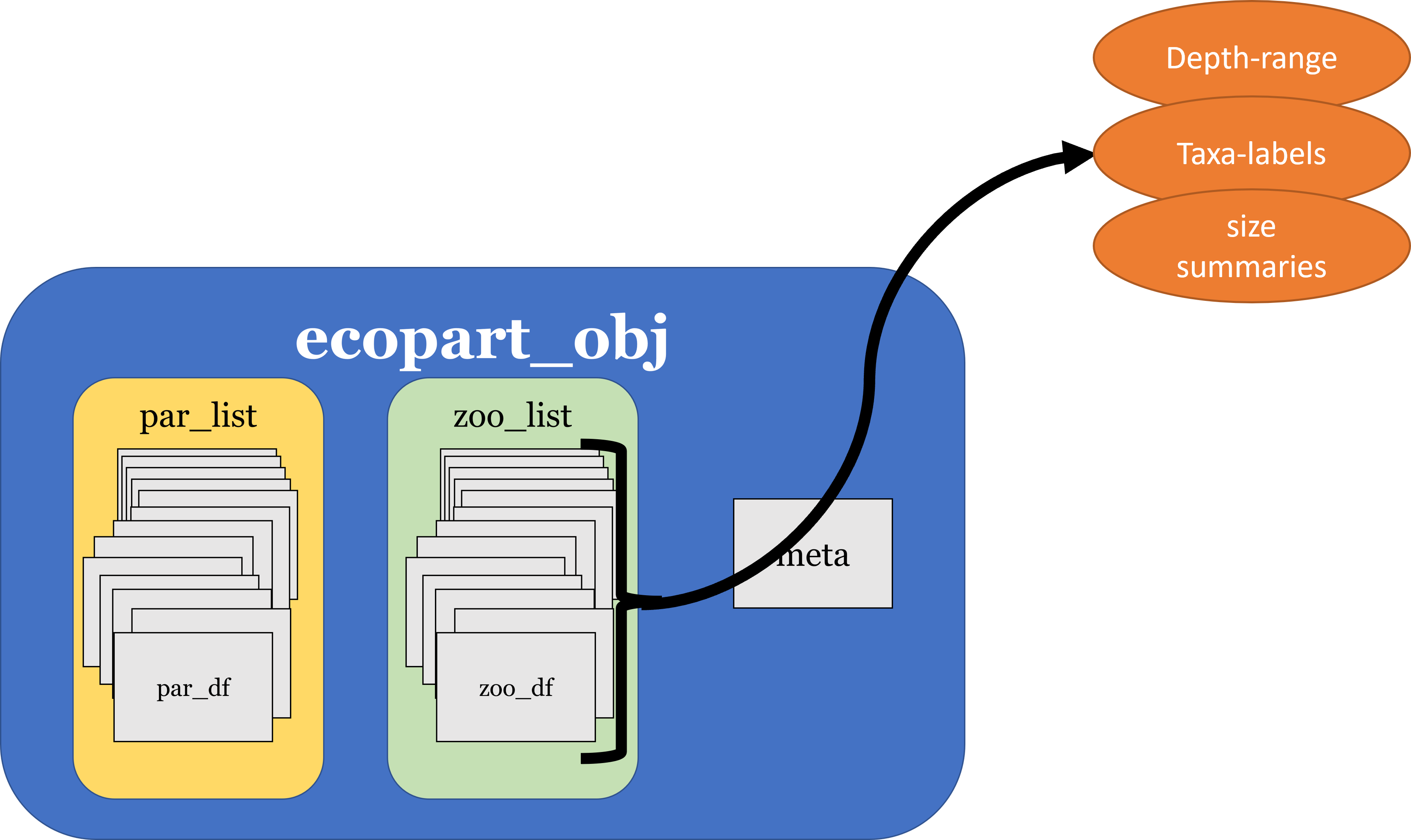
Package tools
Project management:
- Quickly pull information
- Manage metadata
Package tools
Project management:
- Quickly pull information
- Manage metadata
- Relabel categories
Package tools
Project management:
- Quickly pull information
- Manage metadata
- Relabel categories
- Calculating Functions
- Biovolume
Package tools
Summary & Analysis
Package tools
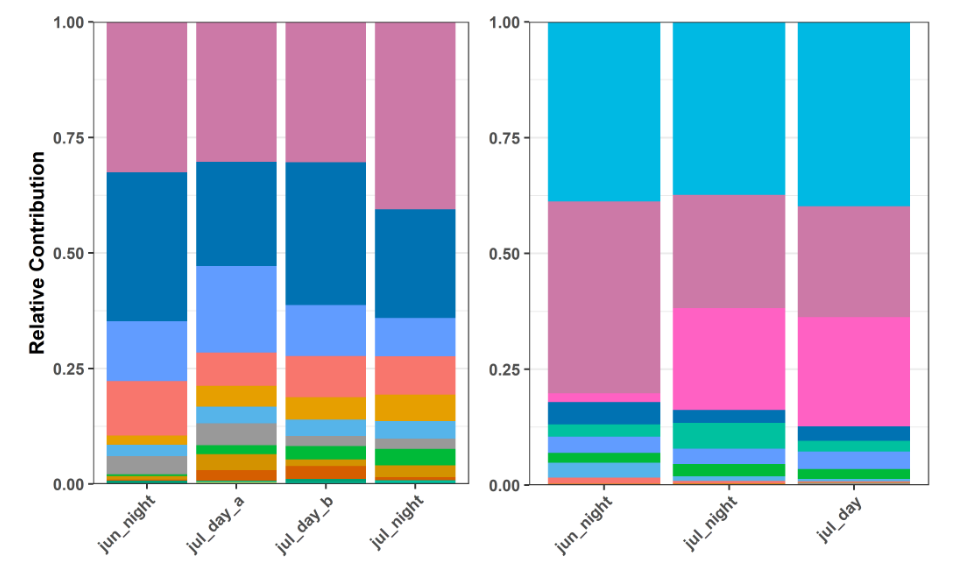
Summary & Analysis
- Relative Taxa
Package tools
Summary & Analysis
- Relative Taxa
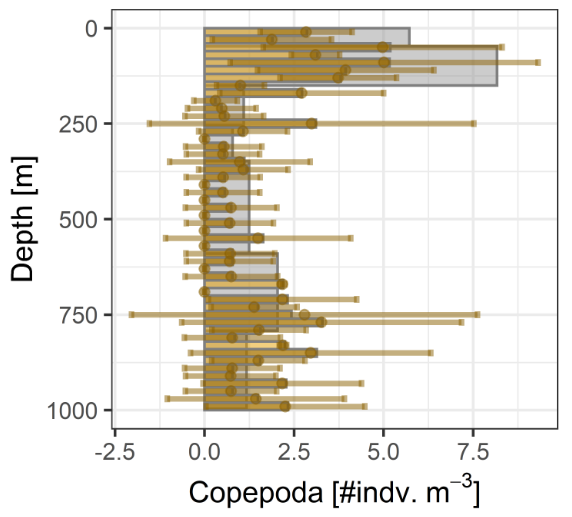
- Calculate Concentrations
Package tools
Summary & Analysis
- Relative Taxa
- Calculate Concentrations
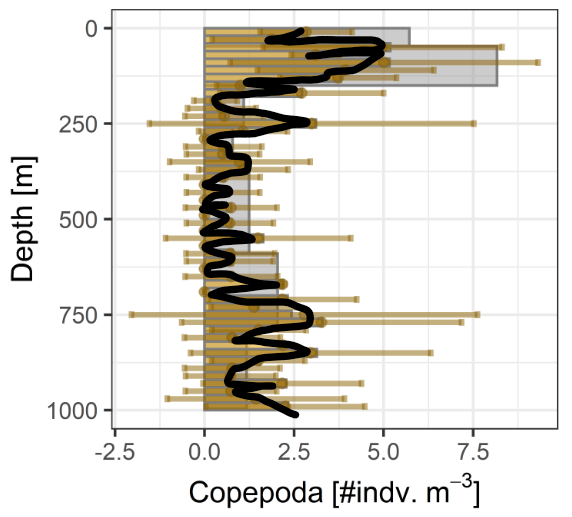
- Averaging Profiles

Package tools
Summary & Analysis
- Relative Taxa
- Calculate Concentrations
- Averaging Profiles
- Depth-integration

Package tools
Summary & Analysis
- Relative Taxa
- Calculate Concentrations
- Averaging Profiles
- Depth-integration
Package tools
Summary & Analysis
- Relative Taxa
- Calculate Concentrations
- Averaging Profiles
- Depth-integration
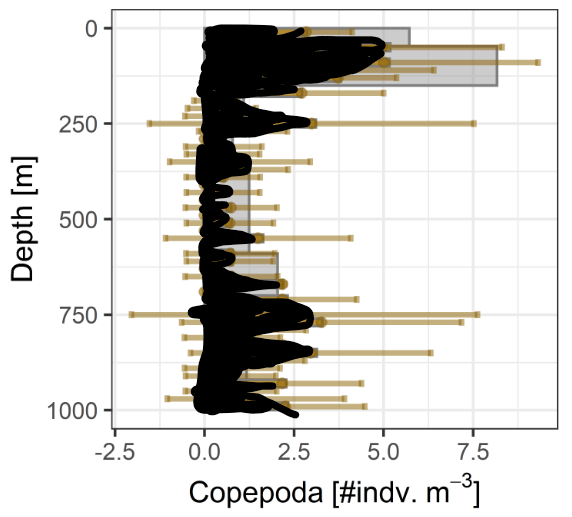
Example Applications
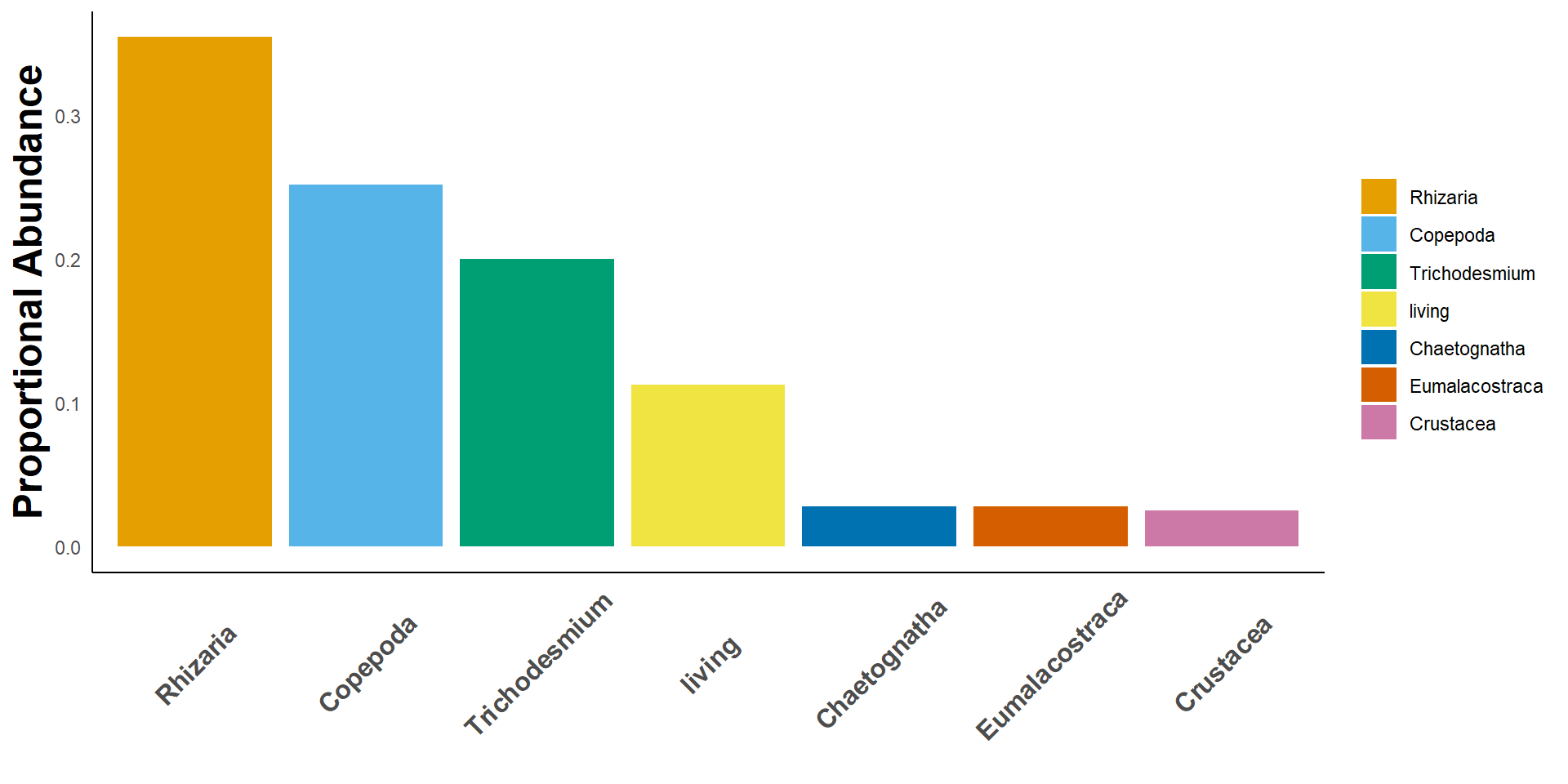
Case 1:
Goal: Get relative abundance of key living taxa
Case 1:
Start with an ecopart_obj
Case 1:
- Drop non-living
Case 1:
- Relabel remaining taxa
Case 1:
- Merge all casts
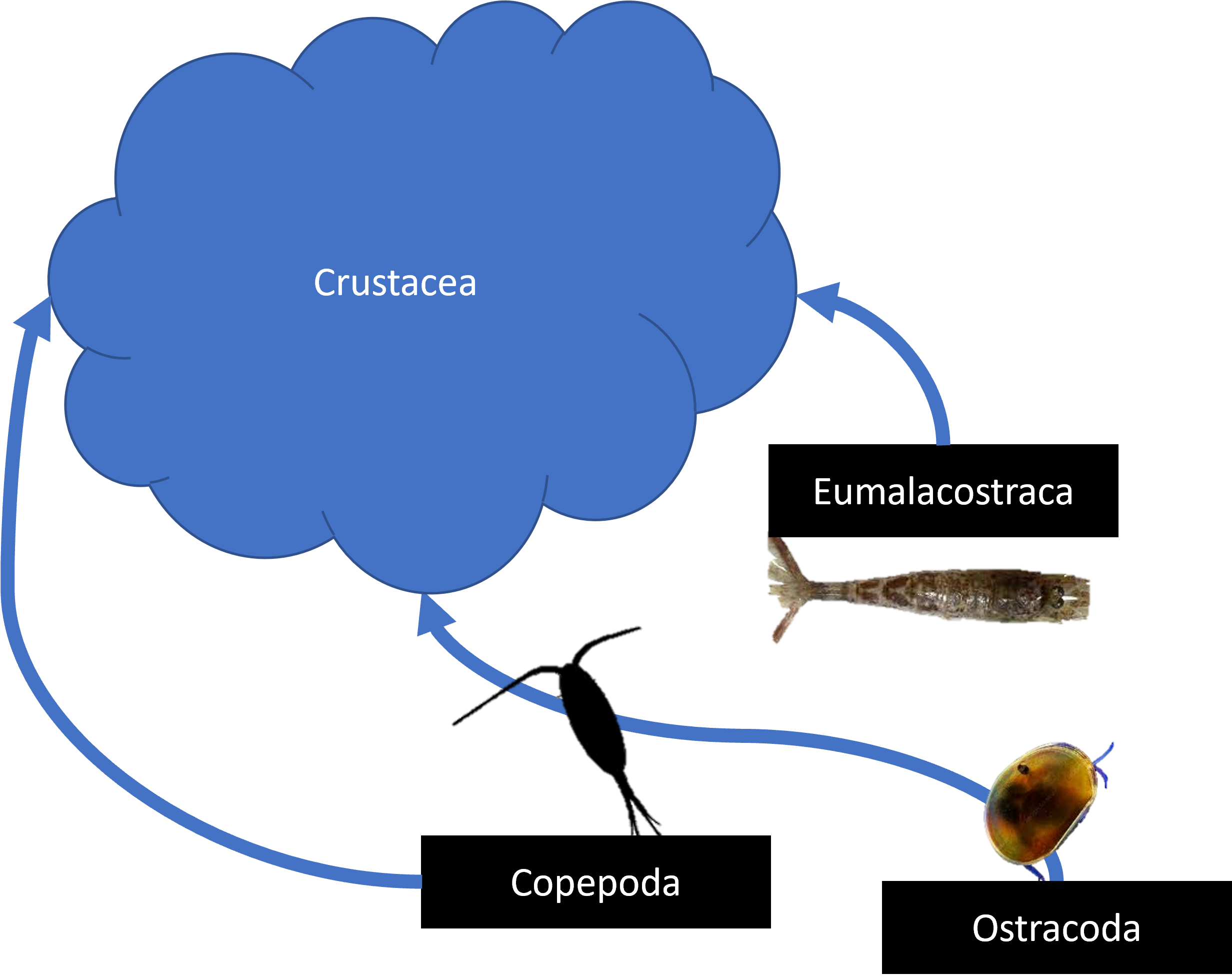
rel_data <- ecopart_example |>
mod_zoo(func = names_drop, drop_names = 'not-living', drop_children = T) |>
add_zoo(func = names_to,
new_names = c('Copepoda', 'Eumalacostraca','Chaetognatha', 'Rhizaria',
'Crustacea','Trichodesmium', 'living'),
suppress_print = T) |>
merge_casts(name_map = list(all_casts = ecopart_example$meta$profileid)) |>Case 1:
- Calculate relative data
rel_data <- ecopart_example |>
mod_zoo(func = names_drop, drop_names = 'not-living', drop_children = T) |>
add_zoo(func = names_to,
new_names = c('Copepoda', 'Eumalacostraca','Chaetognatha', 'Rhizaria',
'Crustacea','Trichodesmium', 'living'),
suppress_print = T) |>
merge_casts(name_map = list(all_casts = ecopart_example$meta$profileid)) |>
rel_taxa()Case 1:
rel_data <- ecopart_example |>
mod_zoo(func = names_drop, drop_names = 'not-living', drop_children = T) |>
add_zoo(func = names_to, col_name = 'name',
new_names = c('Copepoda', 'Eumalacostraca','Chaetognatha', 'Rhizaria',
'Crustacea','Trichodesmium', 'living'),
suppress_print = T) |>
merge_casts(name_map = list(all_casts = ecopart_example$meta$profileid)) |>
rel_taxa()| taxa | rel_abundance |
|---|---|
| Rhizaria | 0.3544304 |
| Copepoda | 0.2516754 |
| Trichodesmium | 0.1999255 |
| living | 0.1124348 |
| Chaetognatha | 0.0282949 |
| Eumalacostraca | 0.0279226 |
| Crustacea | 0.0253165 |
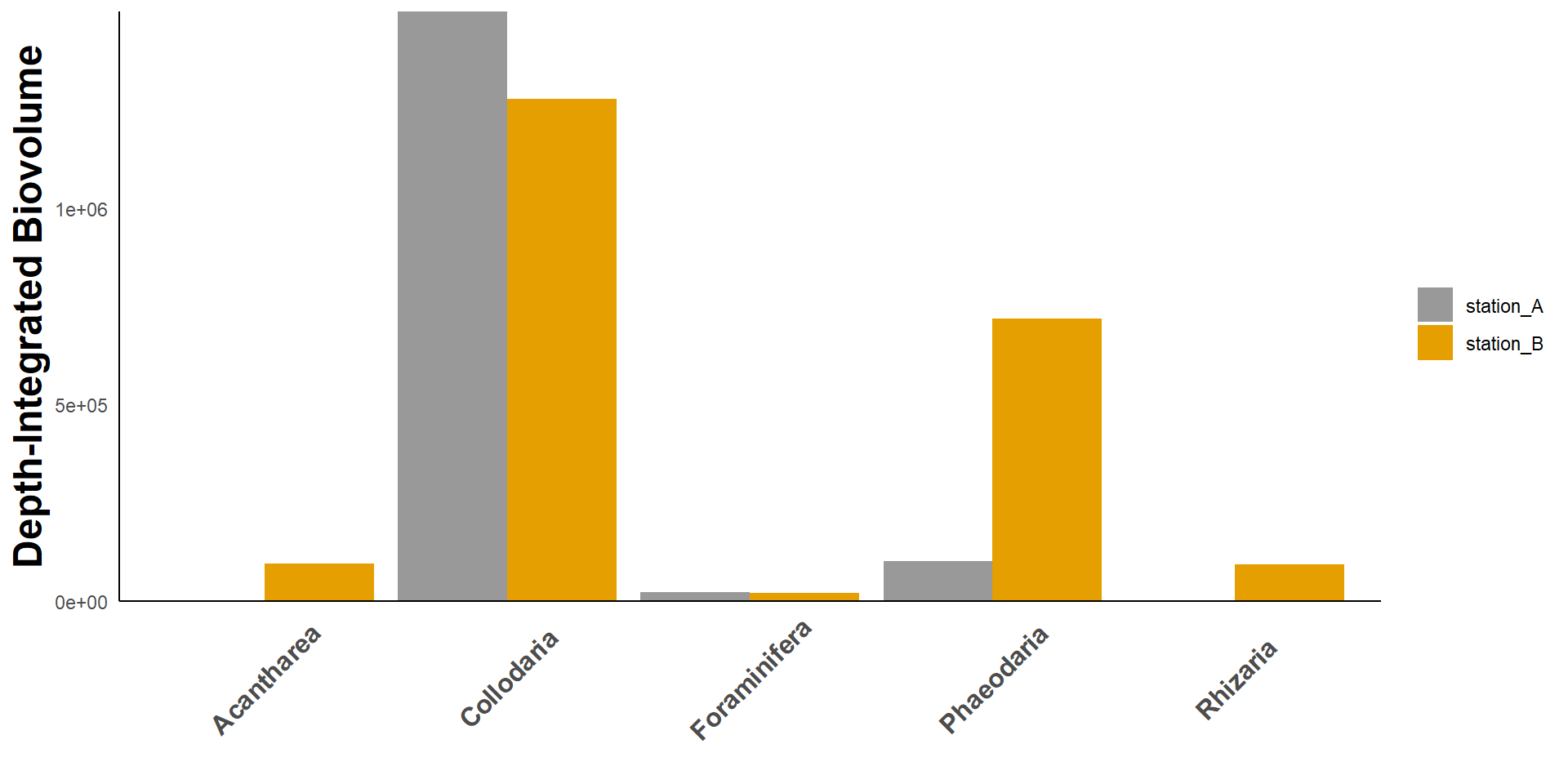
Case 2:
Goal: Calculate depth-integrated biovolume of rhizaria taxa
Case 2:
- Keep only rhizaria
Case 2:
- Relabel to new groups
Case 2:
- Calculate biovolume
integrated_rhiz <- ecopart_example |>
mod_zoo(names_keep, keep_names = 'Rhizaria', keep_children = T) |>
add_zoo(names_to, col_name = 'name',
new_names = c('Phaeodaria','Foraminifera','Acantharea','Rhizaria','Collodaria'),
suppress_print = T) |>
add_zoo(biovolume, col_name = 'biovol', shape = "sphere", pixel_mm = 0.92) |>Case 2:
- Calculate biovolume concentration
integrated_rhiz <- ecopart_example |>
mod_zoo(names_keep, keep_names = 'Rhizaria', keep_children = T) |>
add_zoo(names_to, col_name = 'name',
new_names = c('Phaeodaria','Foraminifera','Acantharea','Rhizaria','Collodaria'),
suppress_print = T) |>
add_zoo(biovolume, col_name = 'biovol', shape = "sphere", pixel_mm = 0.92) |>
uvp_zoo_conc(cast_name = c('bats361_ctd1', 'bats361_ctd3', 'bats361_ctd5',
'bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'),
func_col = 'biovol', func = sum,
breaks = seq(0,1200,25)) |>Case 2:
- Average by station
integrated_rhiz <- ecopart_example |>
mod_zoo(names_keep, keep_names = 'Rhizaria', keep_children = T) |>
add_zoo(names_to, col_name = 'name',
new_names = c('Phaeodaria','Foraminifera','Acantharea','Rhizaria','Collodaria'),
suppress_print = T) |>
add_zoo(biovolume, col_name = 'biovol', shape = "sphere", pixel_mm = 0.92) |>
uvp_zoo_conc(cast_name = c('bats361_ctd1', 'bats361_ctd3', 'bats361_ctd5',
'bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'),
func_col = 'biovol', func = sum,
breaks = seq(0,1200,25)) |>
average_casts(name_map = list(
station_A = c('bats361_ctd1, bats361_ctd3', 'bats361_ctd5'),
station_B = c('bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'))) |>Case 2:
- Integrate casts
integrated_rhiz <- ecopart_example |>
mod_zoo(names_keep, keep_names = 'Rhizaria', keep_children = T) |>
add_zoo(names_to, col_name = 'name',
new_names = c('Phaeodaria','Foraminifera','Acantharea','Rhizaria','Collodaria'),
suppress_print = T) |>
add_zoo(biovolume, col_name = 'biovol', shape = "sphere", pixel_mm = 0.92) |>
uvp_zoo_conc(cast_name = c('bats361_ctd1', 'bats361_ctd3', 'bats361_ctd5',
'bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'),
func_col = 'biovol', func = sum,
breaks = seq(0,1200,25)) |>
average_casts(name_map = list(
station_A = c('bats361_ctd1, bats361_ctd3', 'bats361_ctd5'),
station_B = c('bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'))) |>
lapply(integrate_all, need_format = T, subdivisions=1000L) |>
lapply(intg_to_tib)Case 2:
integrated_rhiz <- ecopart_example |>
mod_zoo(names_keep, keep_names = 'Rhizaria', keep_children = T) |>
add_zoo(names_to, col_name = 'name',
new_names = c('Phaeodaria','Foraminifera','Acantharea','Rhizaria','Collodaria'),
suppress_print = T) |>
add_zoo(biovolume, col_name = 'biovol', shape = "sphere", pixel_mm = 0.92) |>
uvp_zoo_conc(cast_name = c('bats361_ctd1', 'bats361_ctd3', 'bats361_ctd5',
'bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'),
func_col = 'biovol', func = sum,
breaks = seq(0,1200,25)) |>
average_casts(name_map = list(
station_A = c('bats361_ctd1, bats361_ctd3', 'bats361_ctd5'),
station_B = c('bats361_ctd2', 'bats361_ctd4', 'bats361_ctd6'))) |>
lapply(integrate_all, need_format = T, subdivisions=1000L) |>
lapply(intg_to_tib)| taxa | intg | station |
|---|---|---|
| Collodaria | 1499653.99 | station_A |
| Foraminifera | 21861.95 | station_A |
| Phaeodaria | 101105.74 | station_A |
| Acantharea | 0.00 | station_A |
| Rhizaria | 0.00 | station_A |
| Acantharea | 95261.69 | station_B |
| Collodaria | 1278423.93 | station_B |
| Foraminifera | 21019.25 | station_B |
| Phaeodaria | 719243.76 | station_B |
| Rhizaria | 93708.84 | station_B |
See for yourself!
Future Directions:
Future:
- Expand tools & scope

Future:
- Expand tools & scope
- Speed up functions
Future:
- Expand tools & scope
- Speed up functions
- Collaborate
Contact Me:
- AB93@email.sc.edu
- Twitter: @TheAlexBarth
- Web: theAlexBarth.github.io
