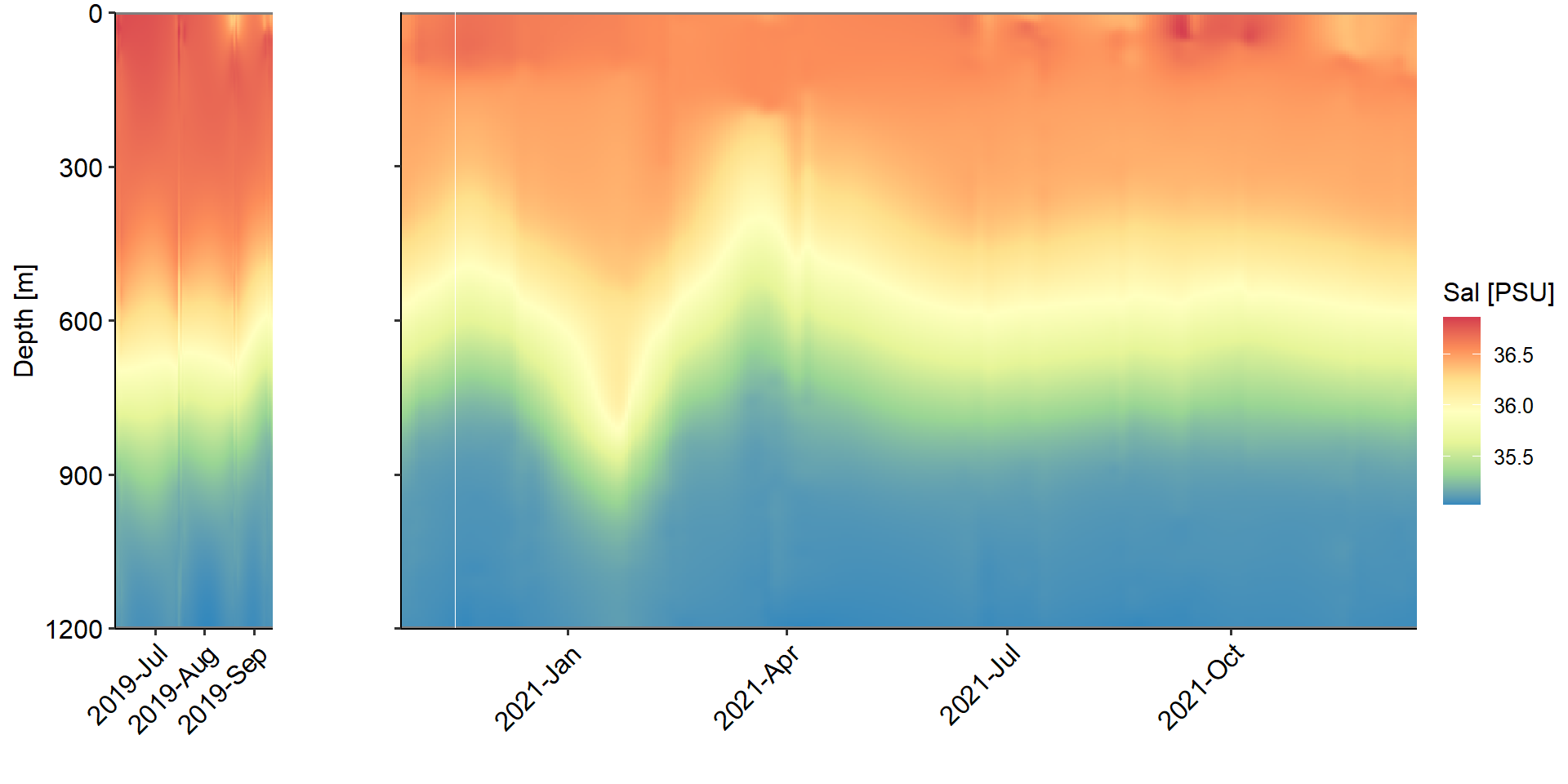
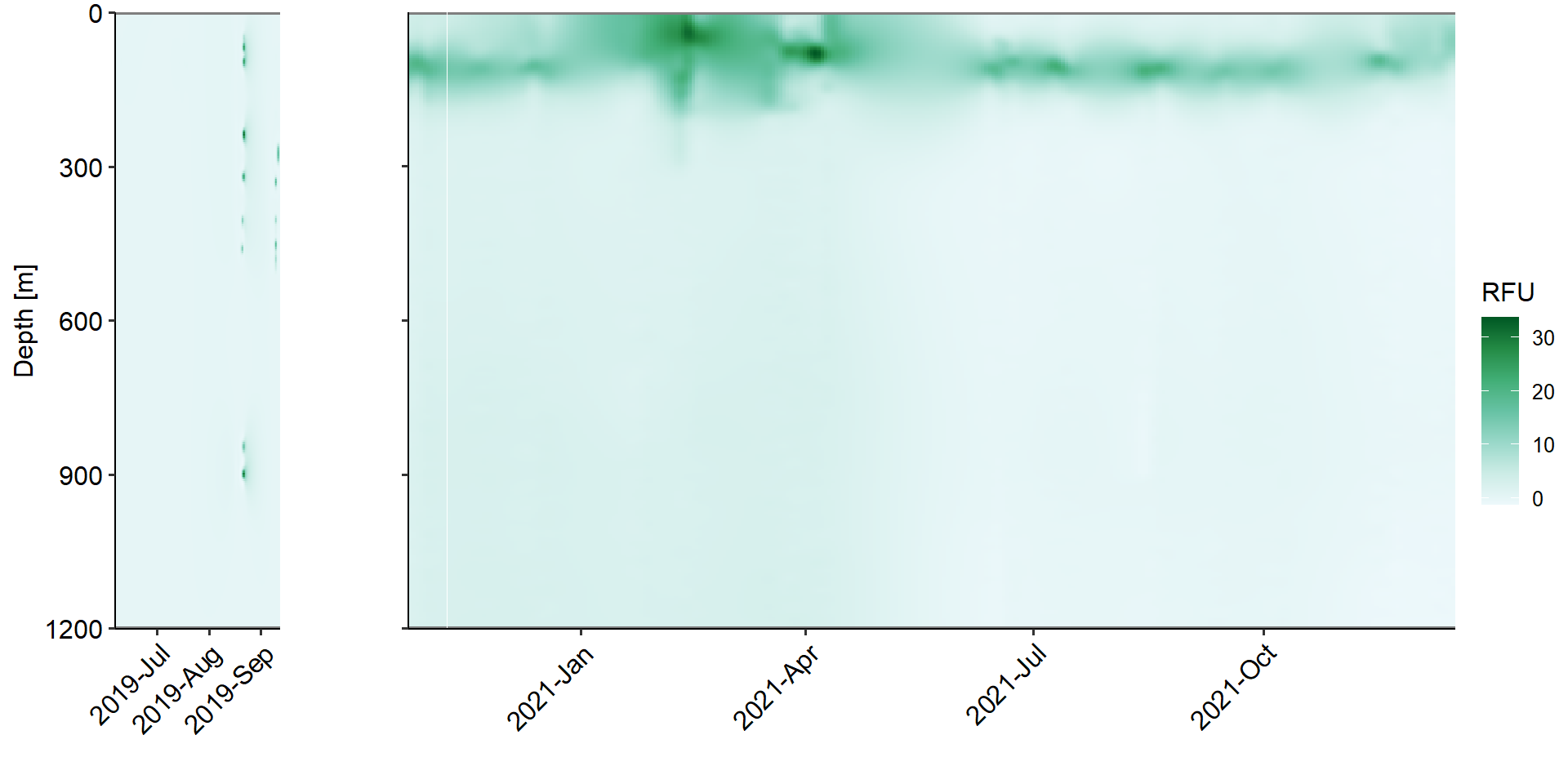
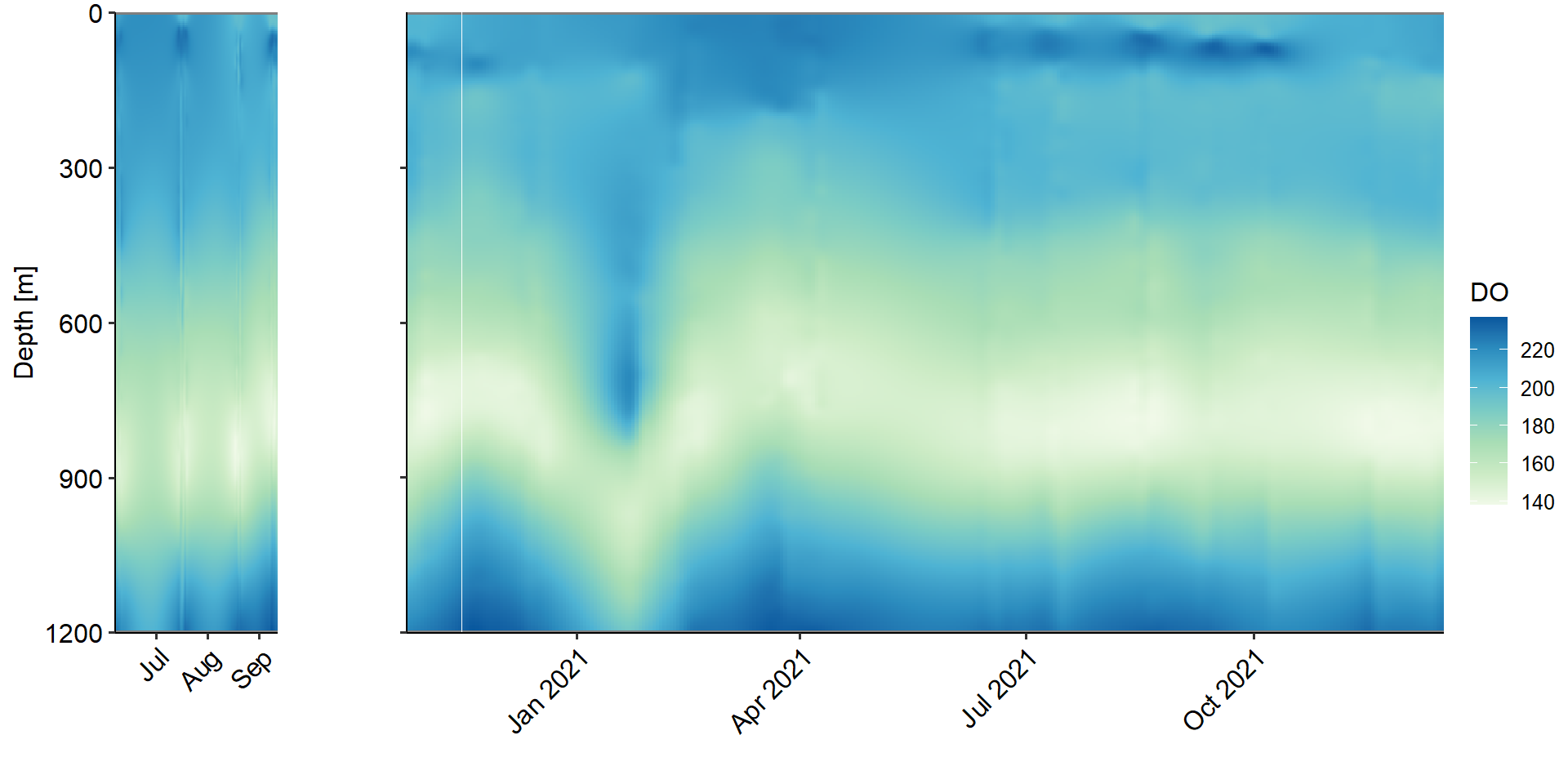
--- title: Physical Backdrop - water column parameters across study period --- ```{r} rm (list = ls ())library (ggplot2)library (ggpubr)<- readRDS ('../data/ctd_02_interp-grid.rds' )$ full_proj<- c ("#feb483" , "#d31f2a" , "#ffc000" , "#27ab19" , "#0db5e6" , "#7139fe" , "#d16cfa" )# set up <- function (cruise){<- vector ('list' ,4 )names (ret_list) <- c ('Sal' ,"Temperature" ,'RFU' ,'DO' )return (ret_list)<- list ()$ early <- value_class ()$ late <- value_class ()# |- Plotting ----------------------- for (i in 1 : length (full_proj_plot)) {$ Sal <- ggplot ()+ geom_tile (data = full_proj_values[[i]]$ Sal,aes (x = Date,y = Depth,fill = Sal)) + scale_y_reverse ()+ scale_fill_distiller (palette = 'Spectral' )+ scale_color_manual (breaks = c ('day' ,'night' ),values = c ('grey' ,'black' ))+ scale_x_datetime (date_labels = '%Y-%b' )+ labs (x = "" ,y = "Depth [m]" , fill = "Sal [PSU]" ,color = "" )+ theme_pubr ()+ theme (legend.position = 'right' )+ coord_cartesian (expand = 0 )$ Temperature <- ggplot ()+ geom_tile (data = full_proj_values[[i]]$ Temperature,aes (x = Date,y = Depth,fill = Temperature)) + scale_y_reverse ()+ scale_x_datetime (date_labels = '%Y-%b' )+ scale_fill_gradientn (colors = rev (ODV_colours))+ scale_color_manual (breaks = c ('day' ,'night' ),values = c ('grey' ,'black' ))+ labs (x = "" ,y = "Depth [m]" , fill = "Temperature" ,color = "" )+ theme_pubr ()+ theme (legend.position = 'right' )+ coord_cartesian (expand = 0 )$ RFU <- ggplot () + geom_tile (data = full_proj_values[[i]]$ RFU,aes (x = Date,y = Depth,fill = RFU)) + scale_y_reverse ()+ scale_x_datetime (date_labels = '%Y-%b' )+ scale_fill_distiller (palette = 'BuGn' ,direction = 1 )+ scale_color_manual (breaks = c ('day' ,'night' ),values = c ('grey' ,'black' ))+ labs (x = "" ,y = "Depth [m]" , fill = "RFU" ,color = "" )+ theme_pubr ()+ theme (legend.position = 'right' )+ coord_cartesian (expand = 0 )$ DO <- ggplot () + geom_tile (data = full_proj_values[[i]]$ DO,aes (x = Date,y = Depth,fill = DO)) + scale_y_reverse ()+ scale_fill_distiller (palette = 'GnBu' ,direction = 1 )+ scale_color_manual (breaks = c ('day' ,'night' ),values = c ('grey' ,'black' ))+ labs (x = "" ,y = "Depth [m]" , fill = "DO" ,color = "" )+ theme_pubr ()+ theme (legend.position = 'right' )+ coord_cartesian (expand = 0 )``` ```{r, fig.cap='Figure 1a. Salinity values across whole study period interpolated from ctd casts.', results='hide',out.width='100%', fig.width=10} #| column: body-outset ggarrange (full_proj_plot$ early$ Sal + theme (axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 )),$ late$ Sal + theme (axis.text.y = element_blank (),axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 ))+ labs (y = "" ),common.legend = T,legend = 'right' ,nrow = 1 ,widths = c (0.2 ,0.8 ),align = 'hv' )``` ```{r, fig.cap='Figure 1b. Temperature values across whole study period interpolated from ctd casts.', results='hide',out.width='100%', fig.width=10} #| column: body-outset ggarrange (full_proj_plot$ early$ Temperature + theme (axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 )),$ late$ Temperature + theme (axis.text.y = element_blank (),axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 ))+ labs (y = "" ),common.legend = T,legend = 'right' ,nrow = 1 ,widths = c (0.2 ,0.8 ),align = 'hv' )``` ```{r, fig.cap='Figure 1c. RFU values across whole study period interpolated from ctd casts.', results='hide',out.width='100%', fig.width=10} #| column: body-outset ggarrange (full_proj_plot$ early$ RFU + theme (axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 )),$ late$ RFU + theme (axis.text.y = element_blank (),axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 ))+ labs (y = "" ),common.legend = T,legend = 'right' ,nrow = 1 ,widths = c (0.2 ,0.8 ),align = 'hv' )``` ```{r, fig.cap='Figure 1d. DO values across whole study period interpolated from ctd casts.', results='hide',out.width='100%', fig.width=10} #| column: body-outset ggarrange (full_proj_plot$ early$ DO + theme (axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 )),$ late$ DO + theme (axis.text.y = element_blank (),axis.text.x = element_text (angle = 45 ,hjust = 1 ,vjust = 1 ))+ labs (y = "" ),common.legend = T,legend = 'right' ,nrow = 1 ,widths = c (0.2 ,0.8 ),align = 'hv' )```