Code
rm(list = ls())
library(cowplot)
library(ggplot2)
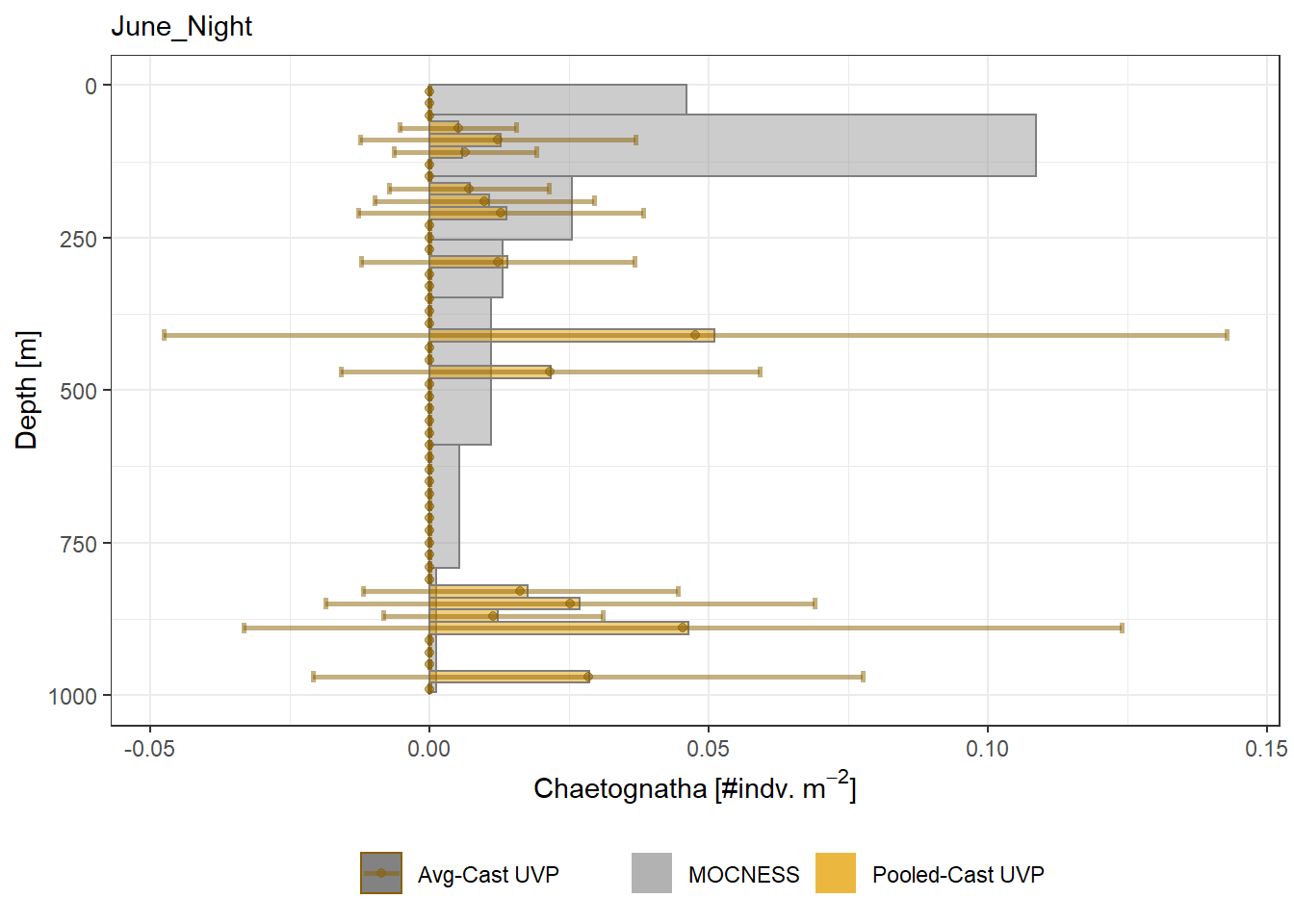
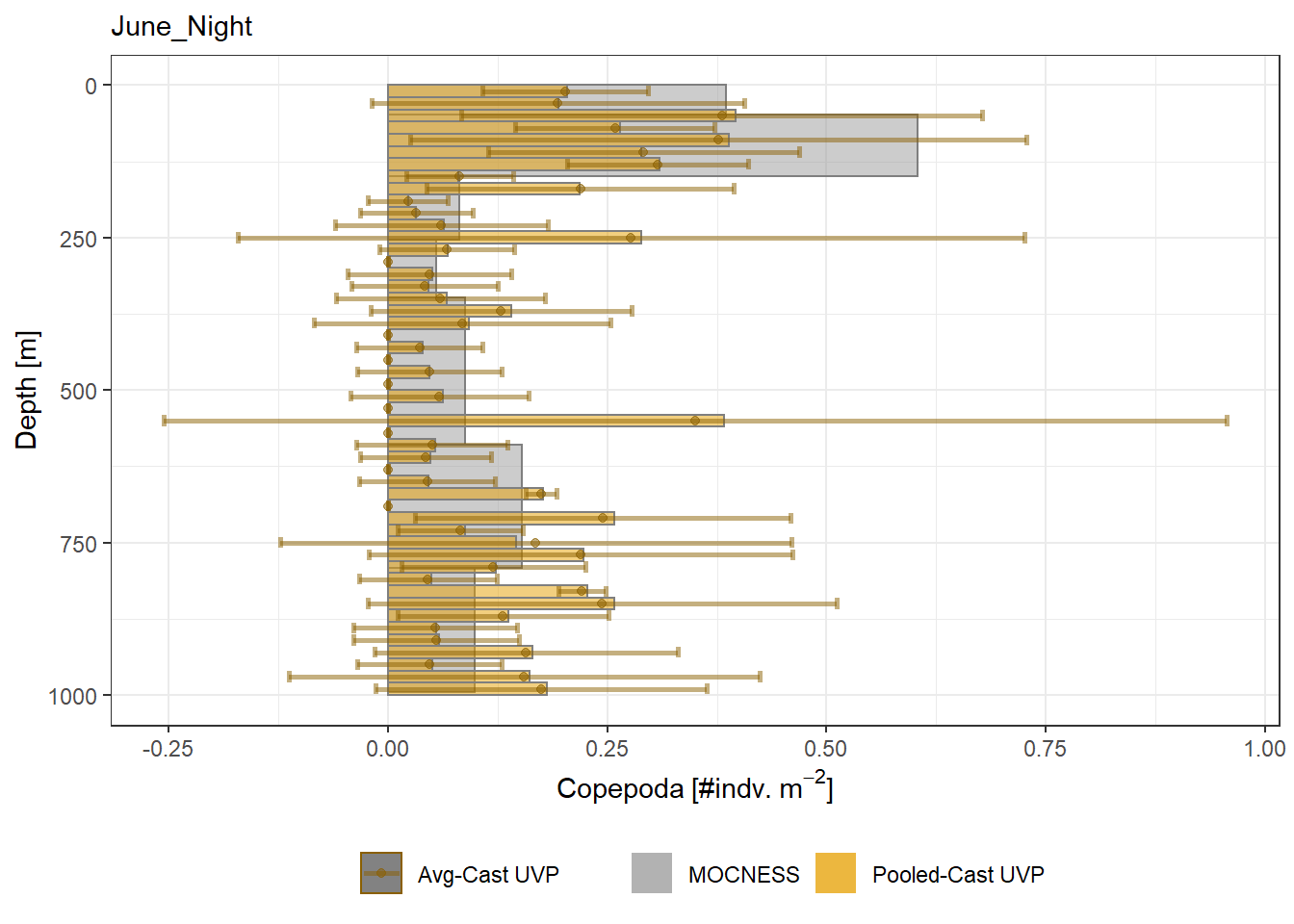
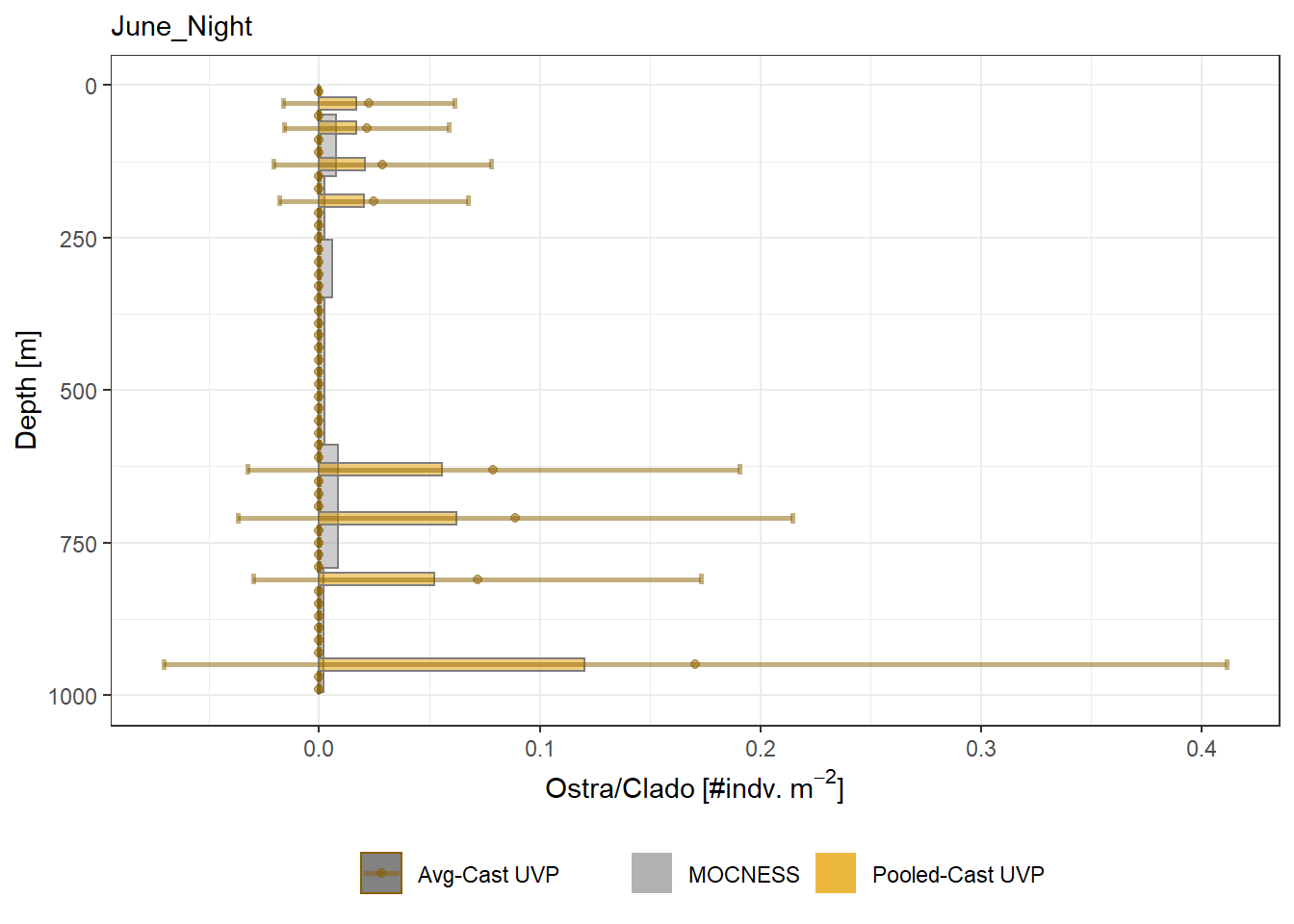
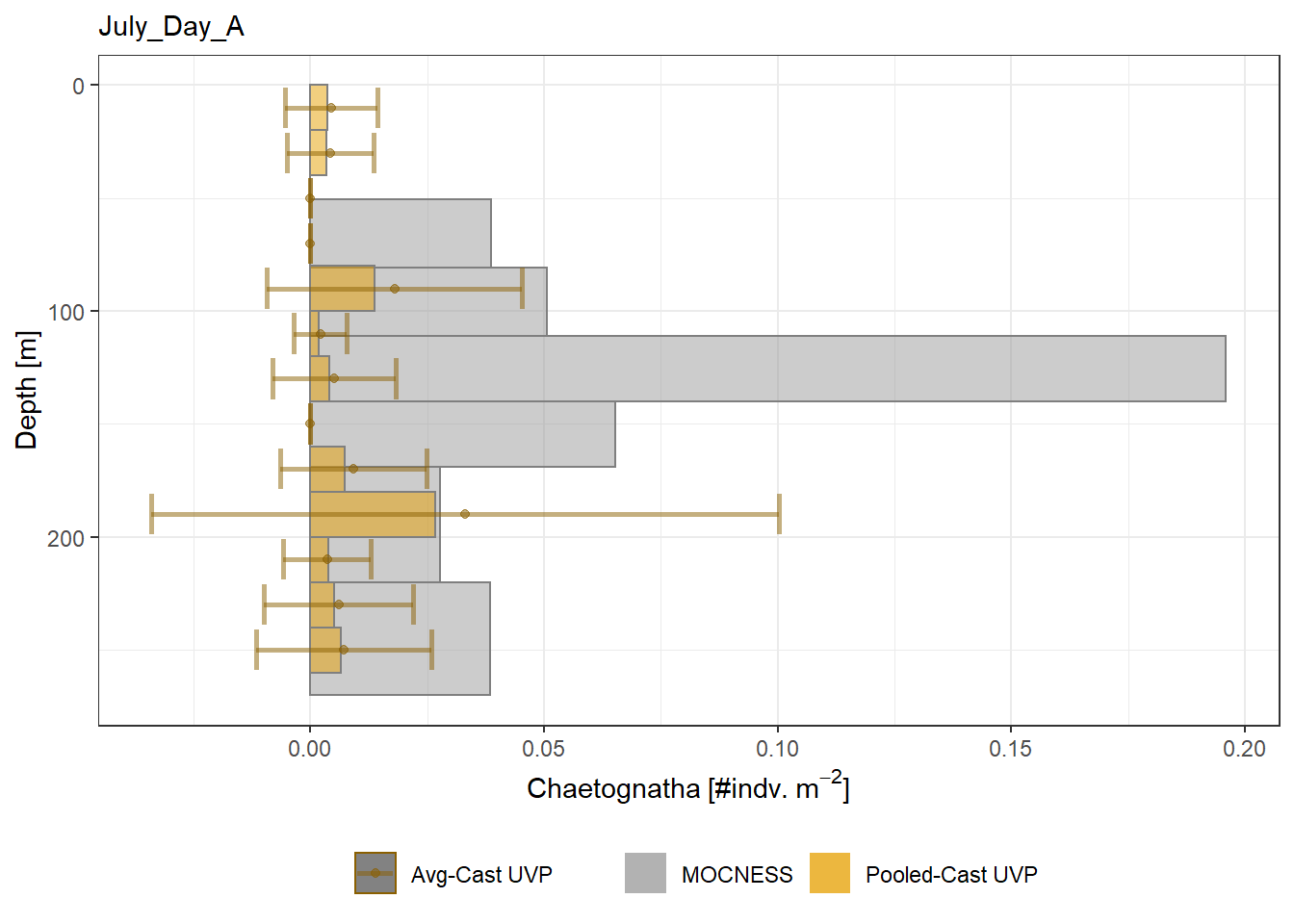
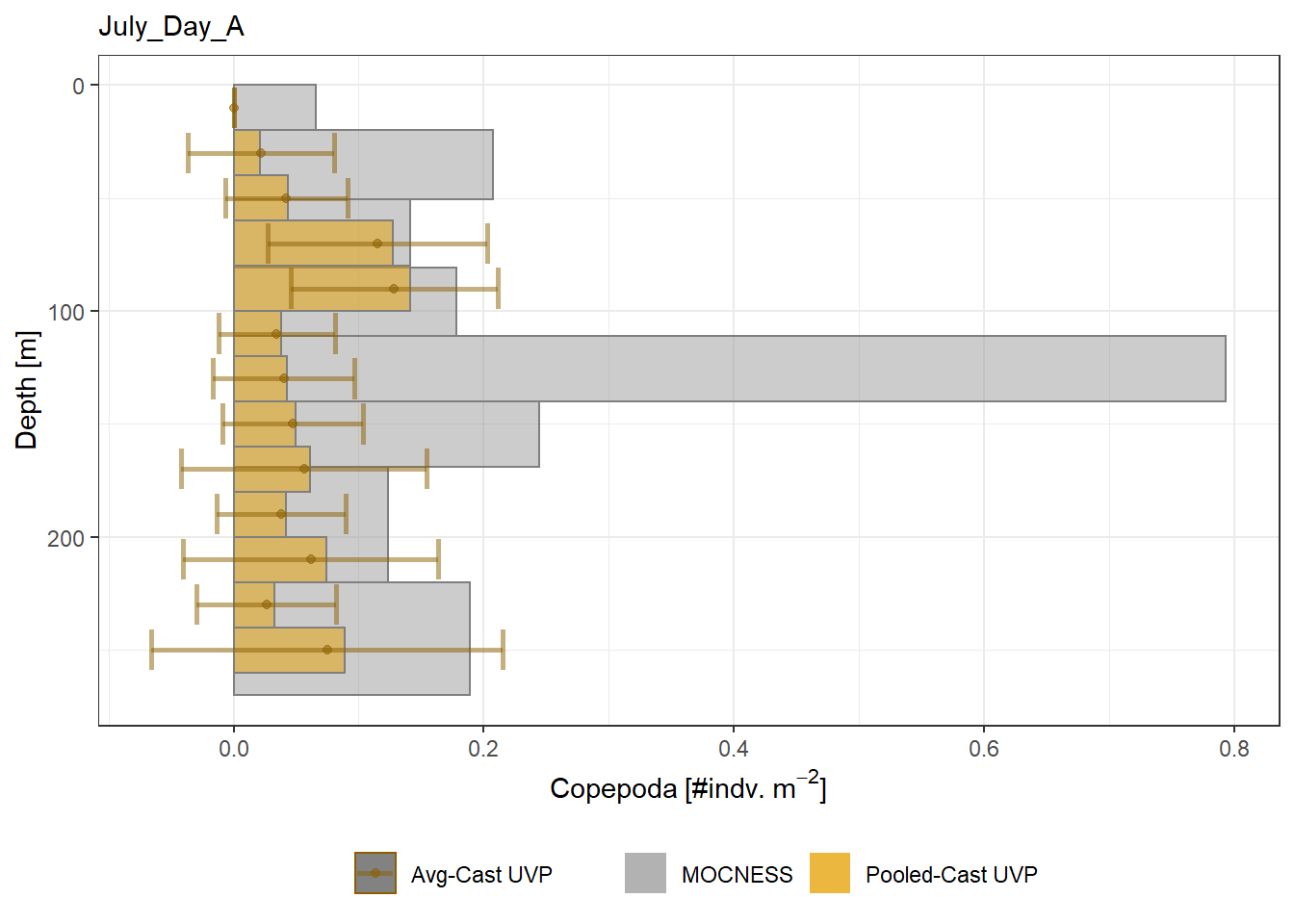
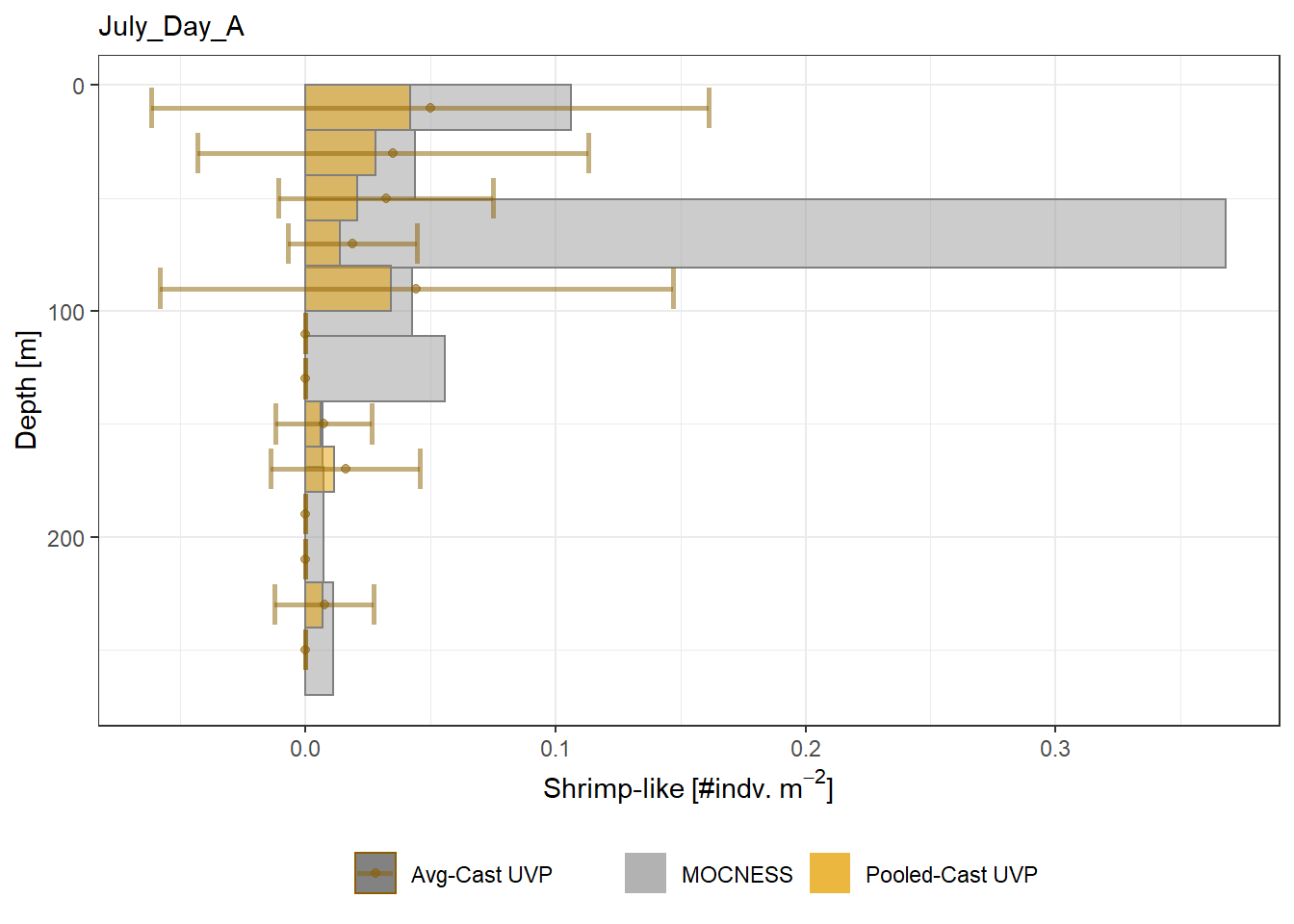
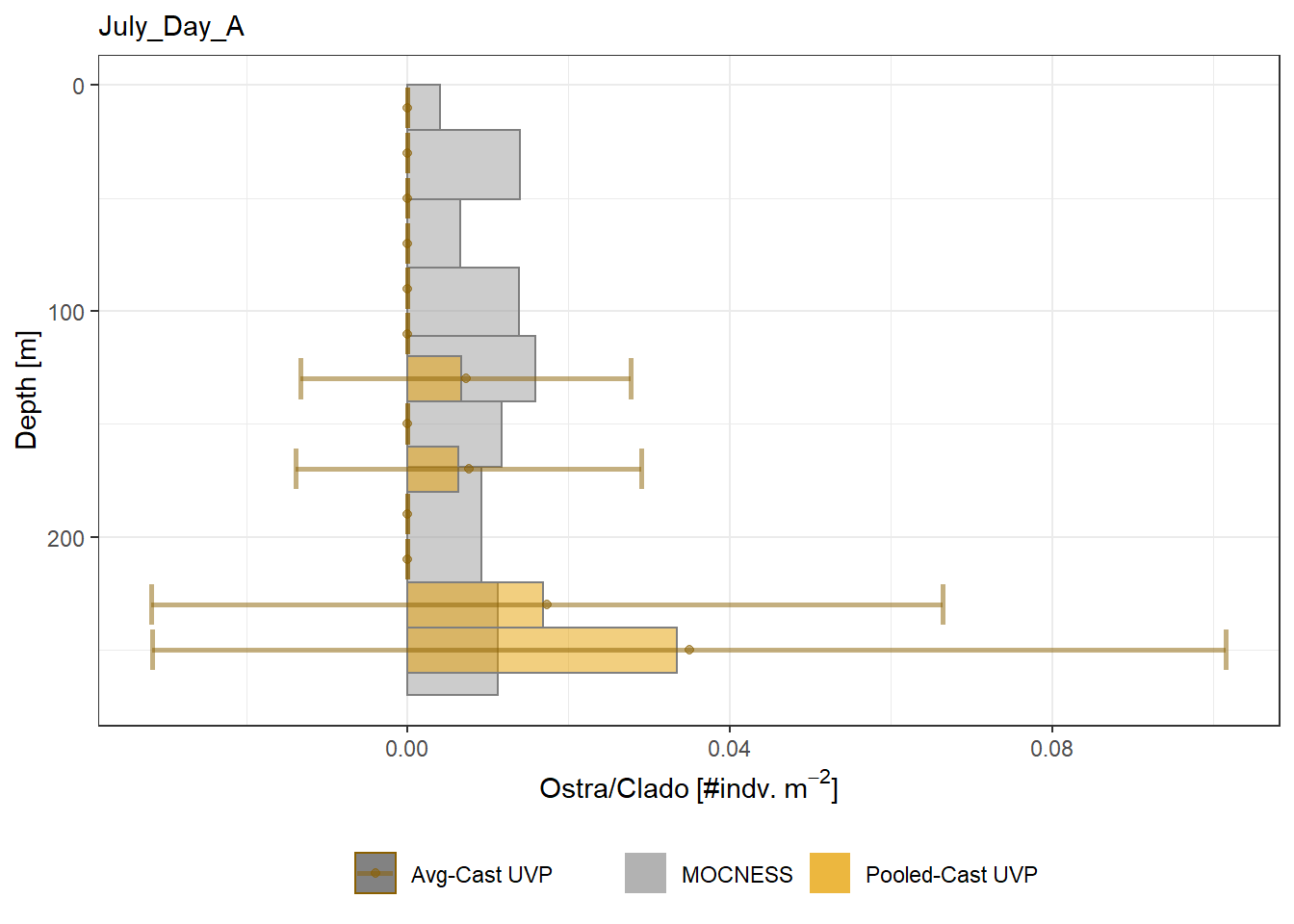
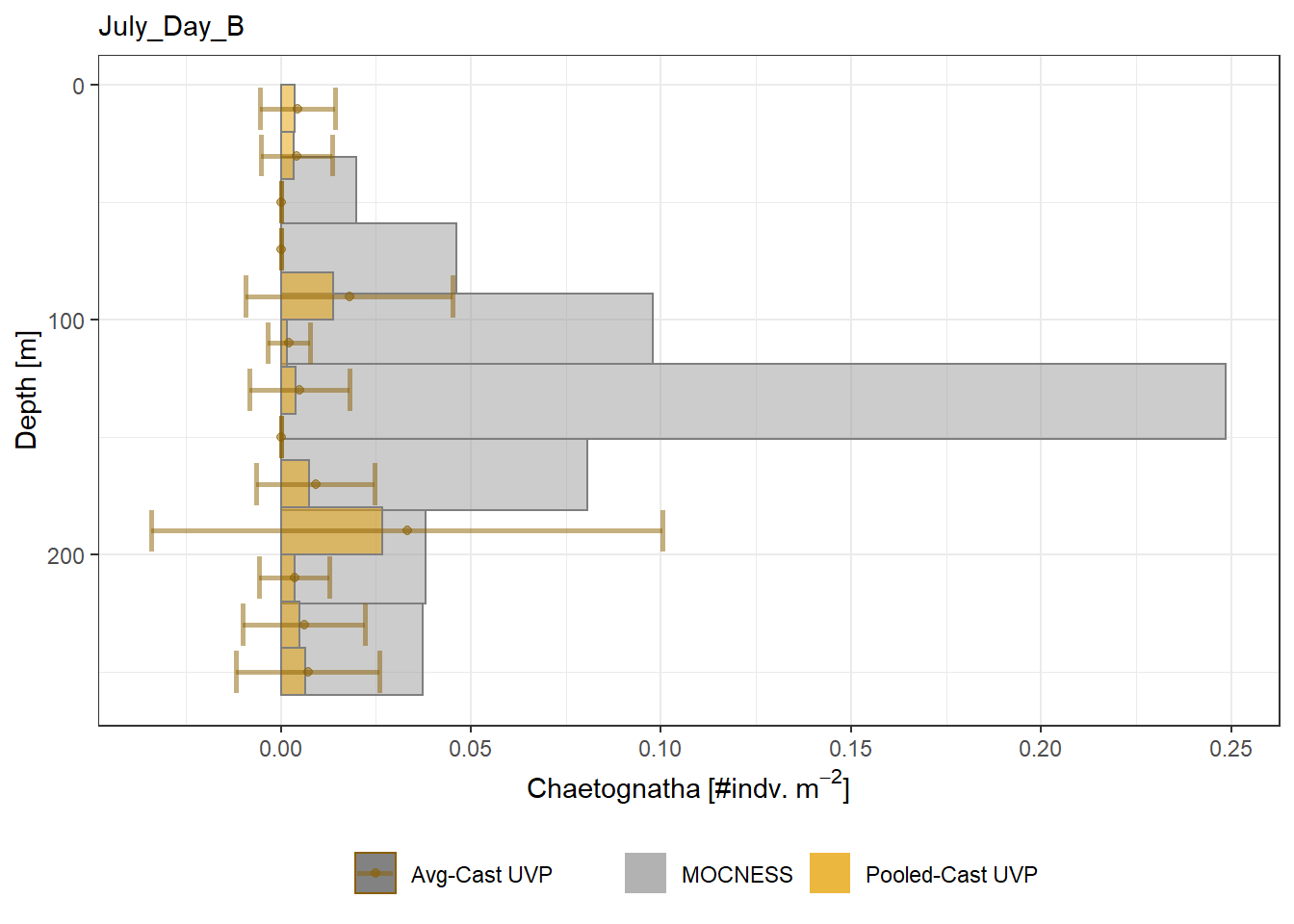
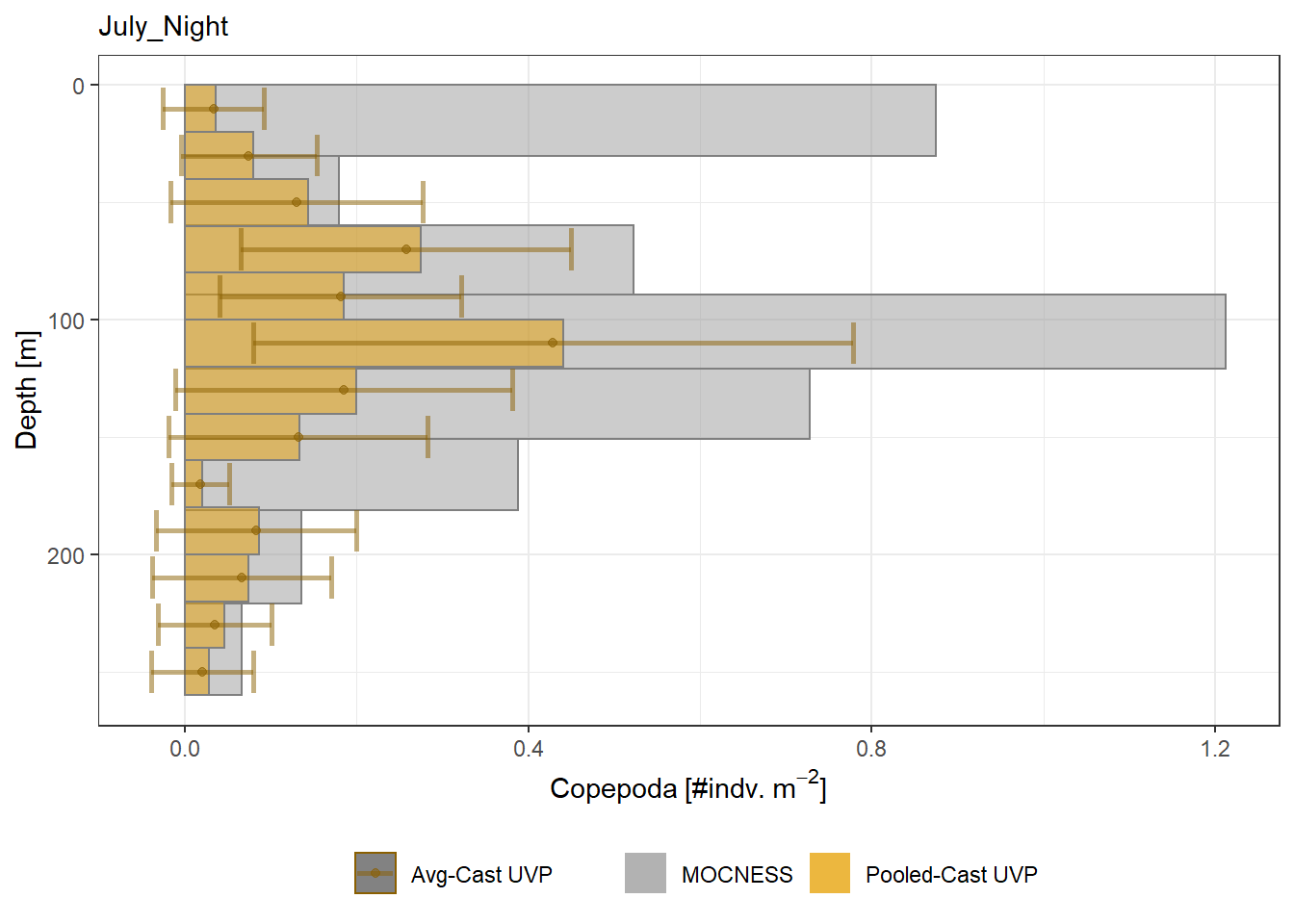
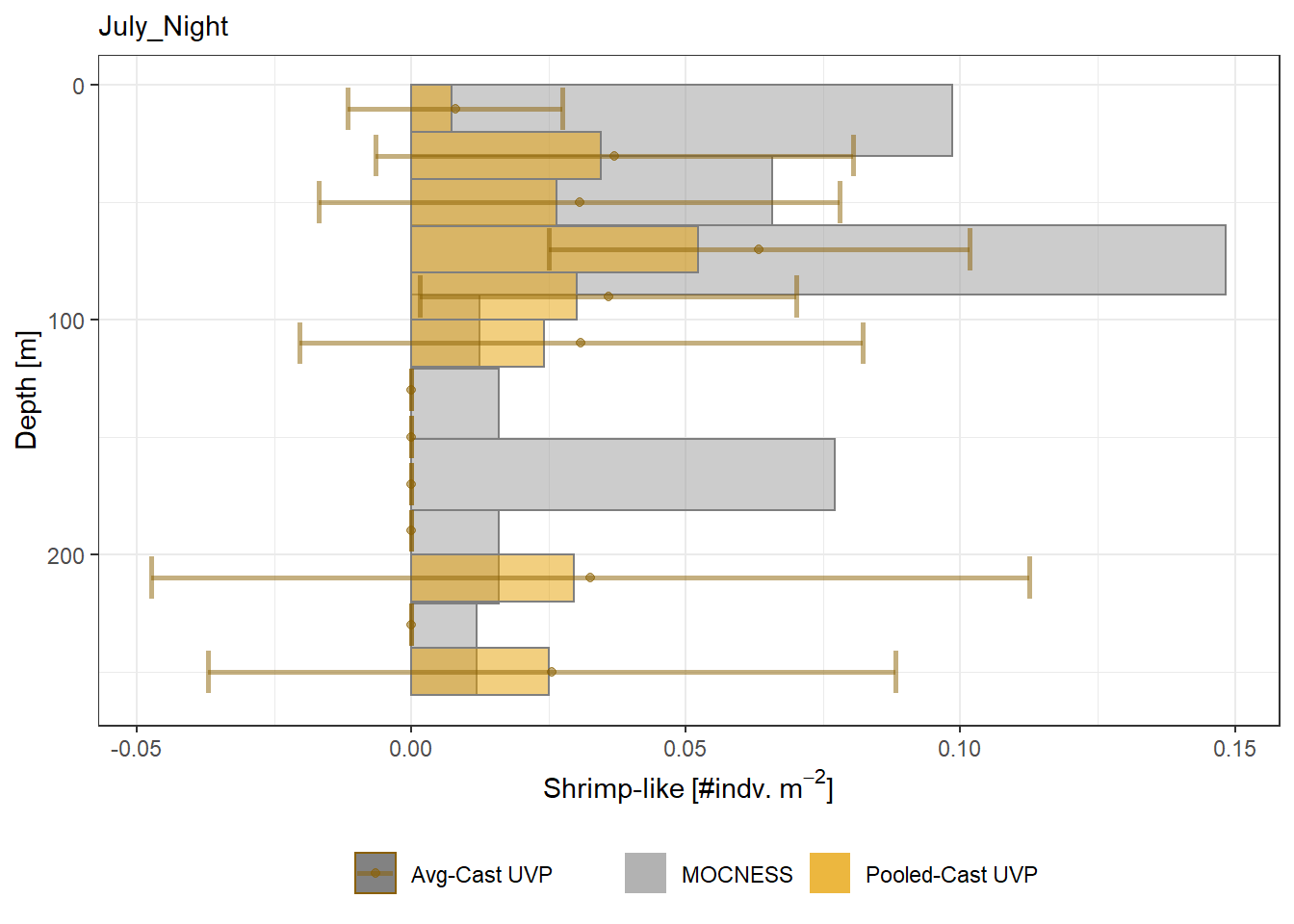
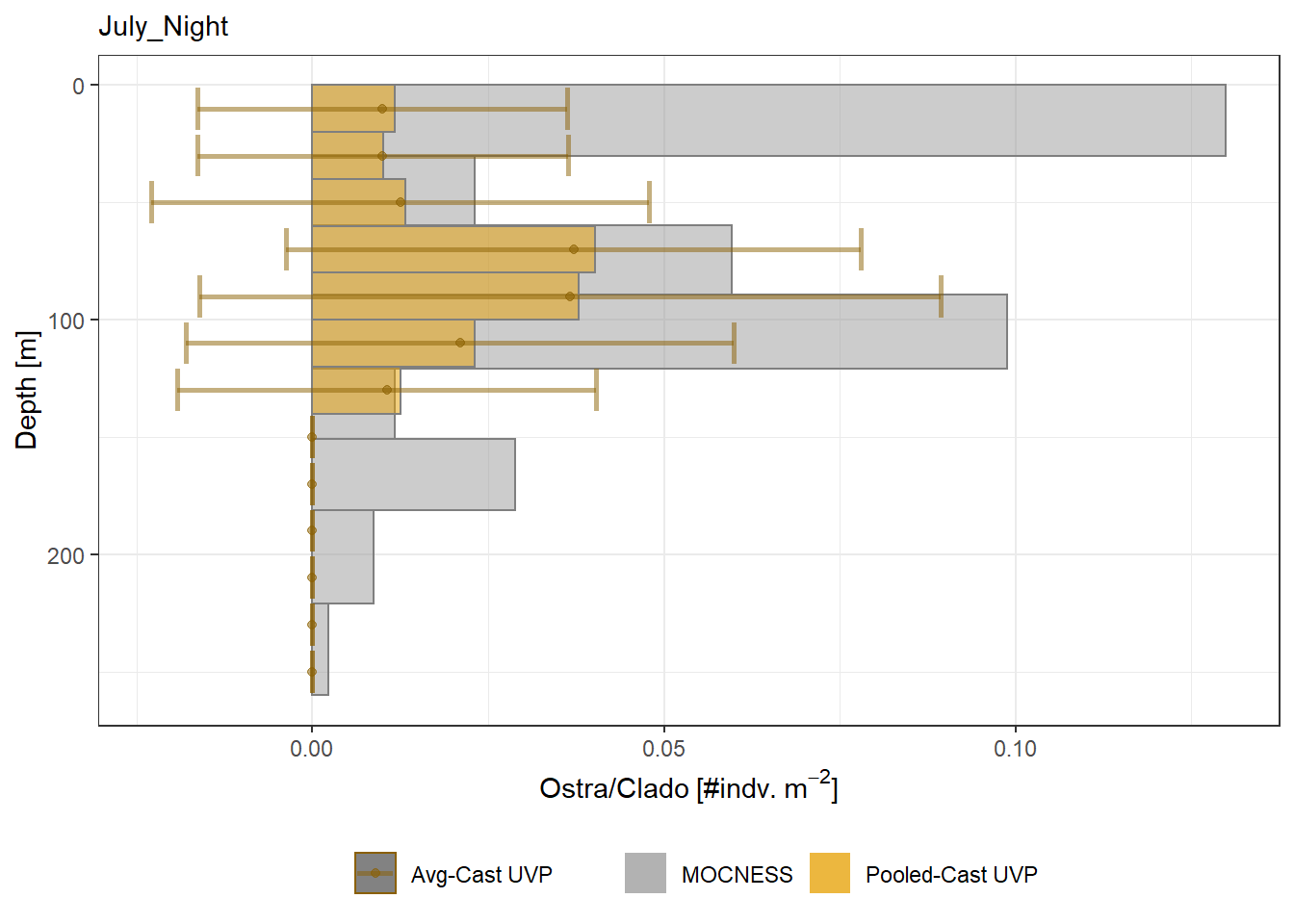
plot_list <- readRDS('../Output/main_fig_06_profile-biomass-comparison.rds')Biomass profiles of comparable taxa are shown when measured by the MOCNESS/ZOOSCAN and the UVP. UVP estimates were calculated two ways. In one method, similar UVP casts are pooled then the biomass concentration of organisms in a depth bin were calculated \[\frac{\sum_{i}^{N}biomass_{i}}{\sum_{i}^{N}volume sampled_{i}}\]. The other method, density is calculated in individual uvp casts, then averaged between all similar casts: \[\frac{\sum_{i=1}^{N} \frac{biomass_{i}}{volume sampled_{i}}}{N}\] For all i casts, with a total of N casts.


---
title: "Supplemental Figure 7 - All Biomass Profiles"
---
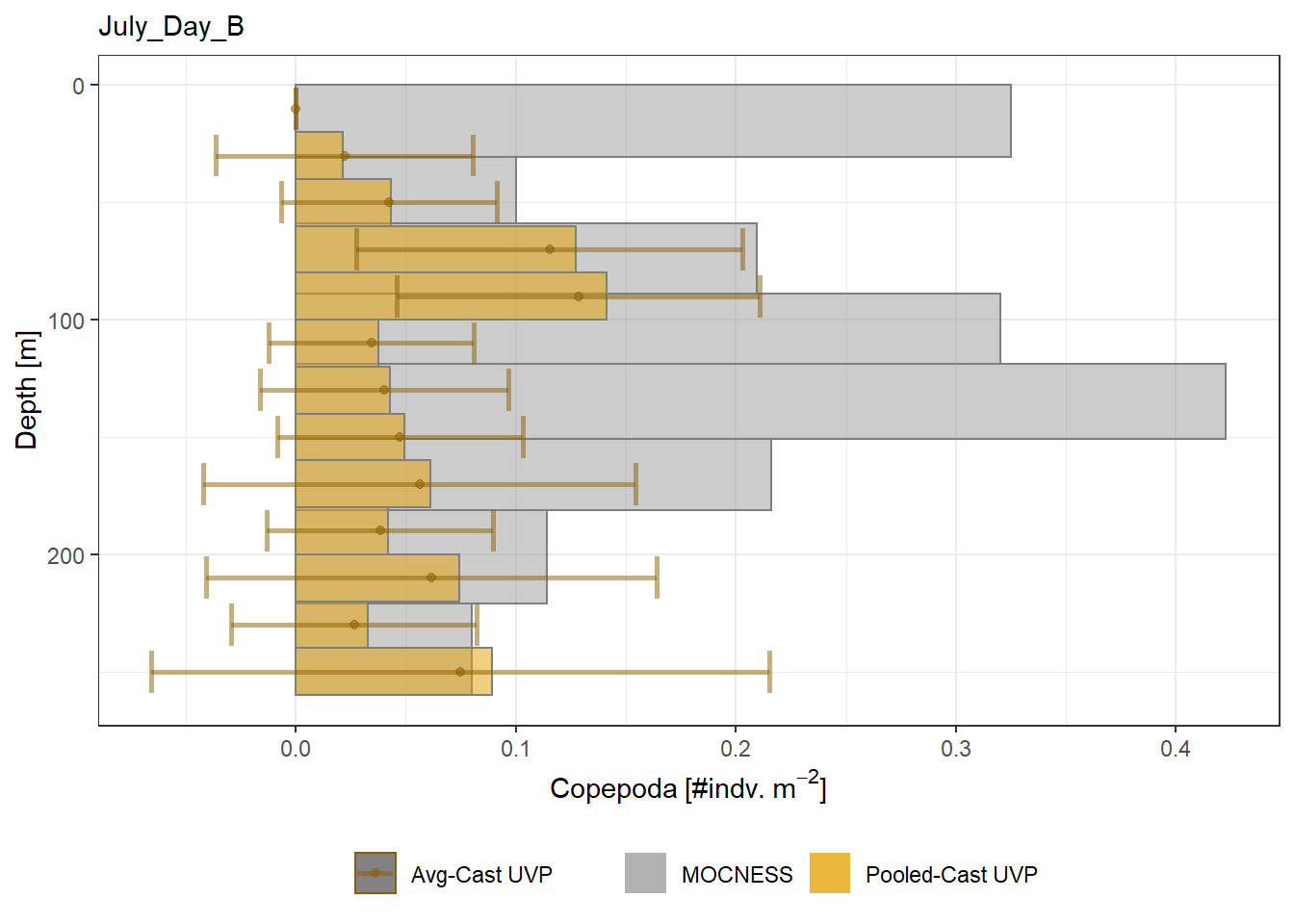
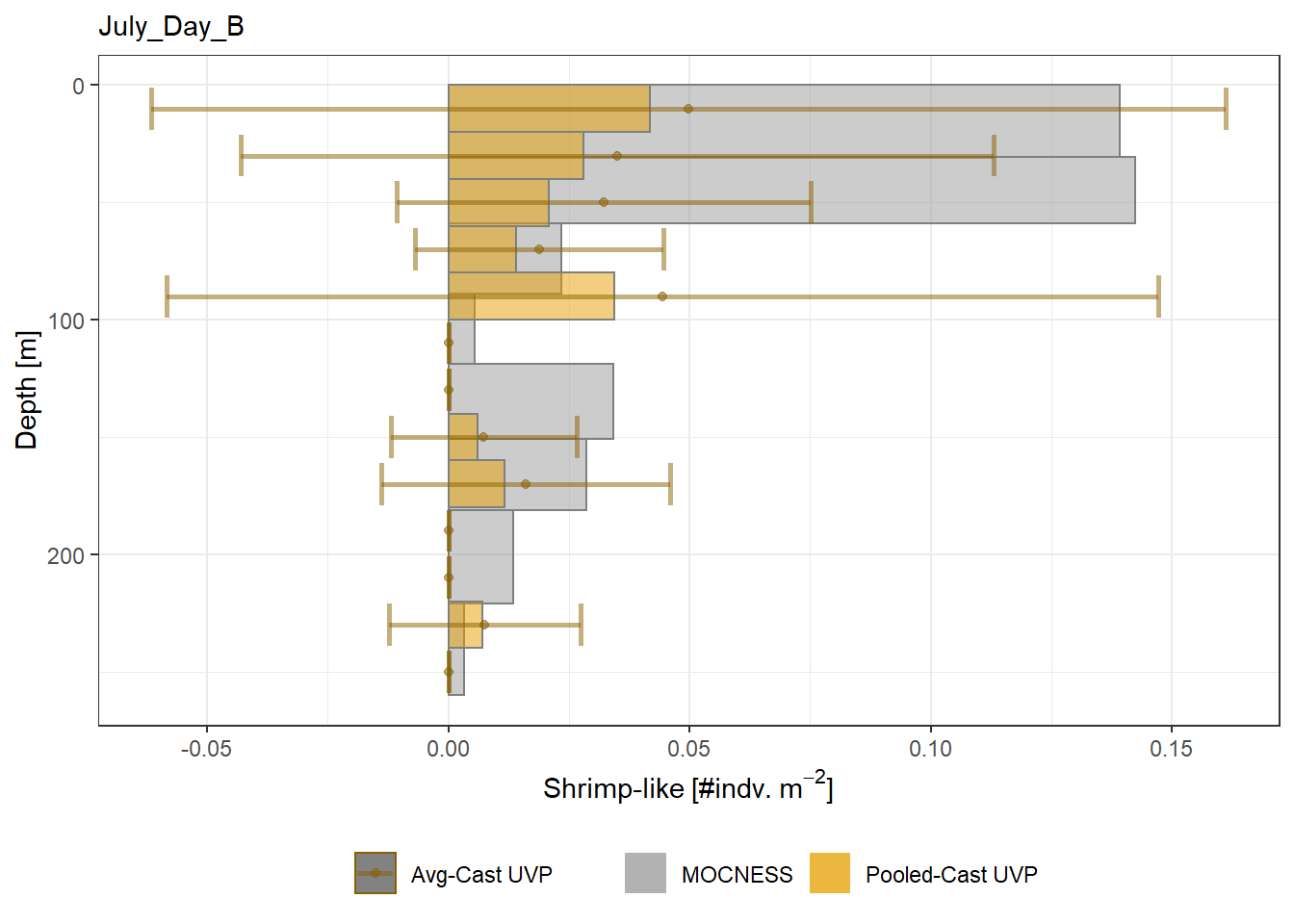
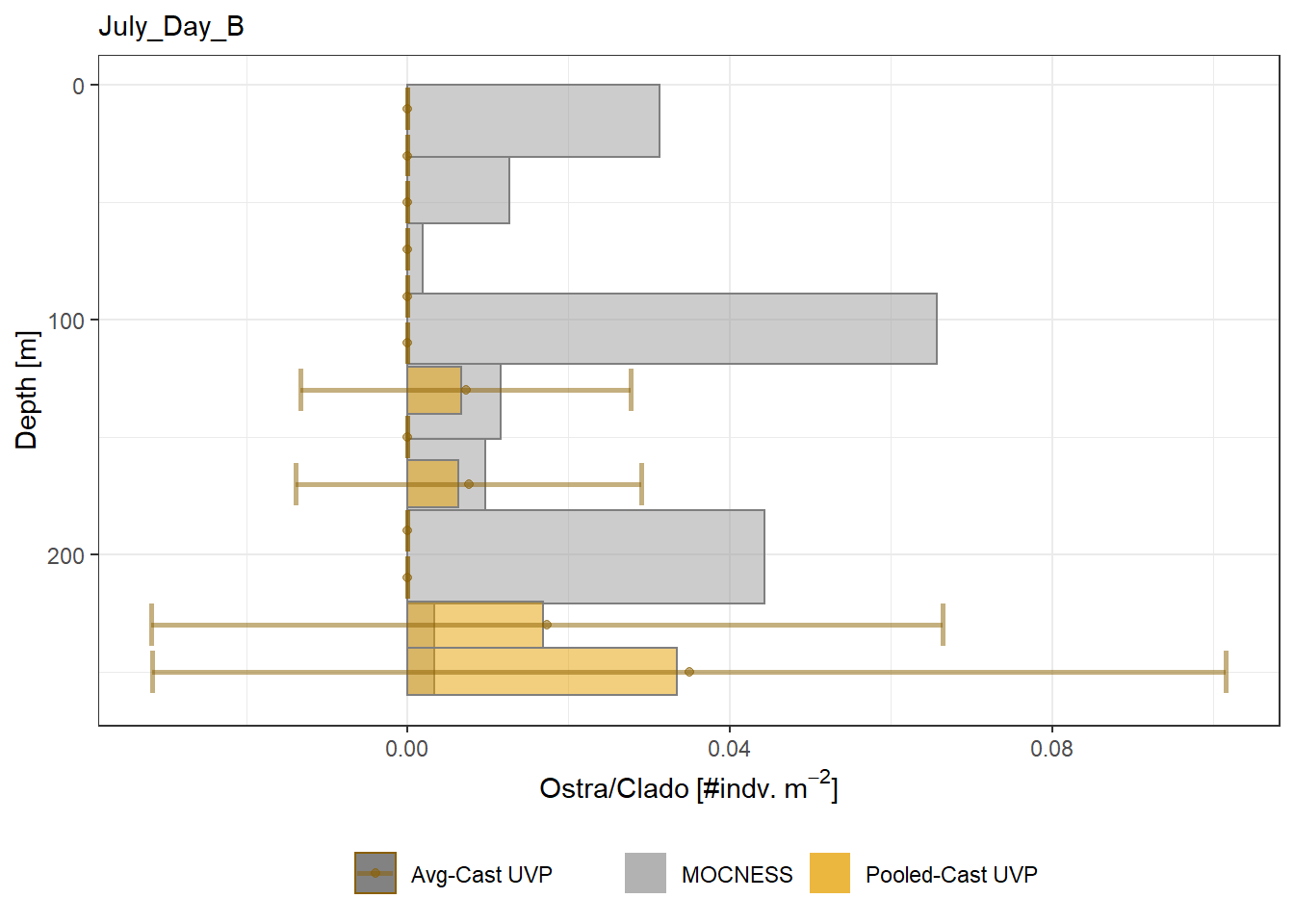
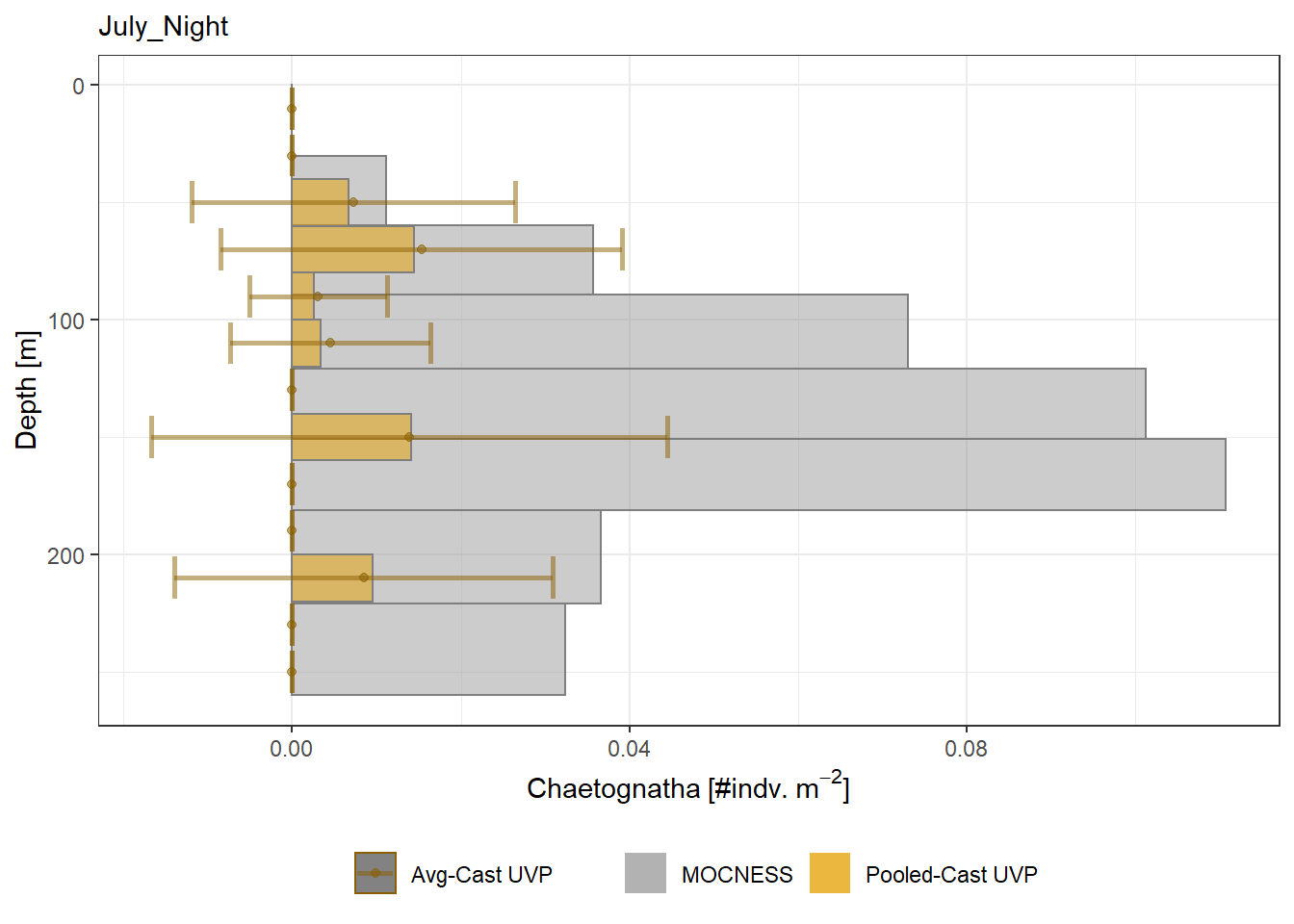
Biomass profiles of comparable taxa are shown when measured by the MOCNESS/ZOOSCAN and the UVP.
UVP estimates were calculated two ways. In one method, similar UVP casts are pooled then the biomass concentration of organisms in a depth bin were calculated $$\frac{\sum_{i}^{N}biomass_{i}}{\sum_{i}^{N}volume sampled_{i}}$$. The other method, density is calculated in individual uvp casts, then averaged between all similar casts: $$\frac{\sum_{i=1}^{N} \frac{biomass_{i}}{volume sampled_{i}}}{N}$$ For all i casts, with a total of N casts.
```{r}
rm(list = ls())
library(cowplot)
library(ggplot2)
plot_list <- readRDS('../Output/main_fig_06_profile-biomass-comparison.rds')
```
```{r All-Plots, fig.cap='Supplemental Figure 7. Biomass Concentration profiles of all taxa. Pooled-UVP cast estimates are shown as barplots. Averaged-UVP casts estimates are shown by points with standard deviation shown.'}
#| column: body-outset
plot_grid(for(i in 1:length(plot_list)){print(plot_list[[i]] + theme(legend.position = 'bottom'))}, ncol = 3)
```